Contextual inquiry in science labs
2015 - 2016 | Novartis | Basel, Switzerland
How do we bring the simplest, most useful collaboration experience to every Novartis scientist working in drug discovery?
Approach
At Novartis, leadership's key challenge was to improve collaboration among specialized, yet isolated, research teams to accelerate drug discovery. As a senior UX designer, my task was to identify the barriers to effective information sharing and create tools that streamlined collaboration, enabling scientists to work more effectively and efficiently.
Please note: I worked on researching and designing for many scientific tools at Novartis, and most of it was under an NDA; this case study has some blurred content
Research
To uncover the root causes of collaboration issues, I conducted research that included:
Stakeholder Interviews: I conducted in-depth interviews with key stakeholders, including scientists and lab managers, to understand their daily workflows, pain points, and needs for collaboration. These interviews revealed that teams often worked in isolation and lacked visibility into each other’s work, which slowed down work.
Surveys: I developed and distributed company-wide surveys targeting a broader group of scientists to gather quantitative data on collaboration habits, tools used, and barriers they faced. The survey responses showed that many felt the current tools were not fit for purpose and that discovering archived knowledge was particularly challenging.
Contextual inquiry in labs: To gain a deeper understanding of the scientists' day-to-day work, I conducted contextual inquiries directly within the labs. By observing the workflows, tools, and interactions in real-time, I identified specific pain points related to collaboration and information exchange that were not evident from interviews alone.
Workshops and observation: I facilitated cross-team workshops where scientists and stakeholders discussed existing collaboration processes, tools, and barriers. Observing these discussions helped me identify the competitive culture between teams, which often discouraged open knowledge-sharing.
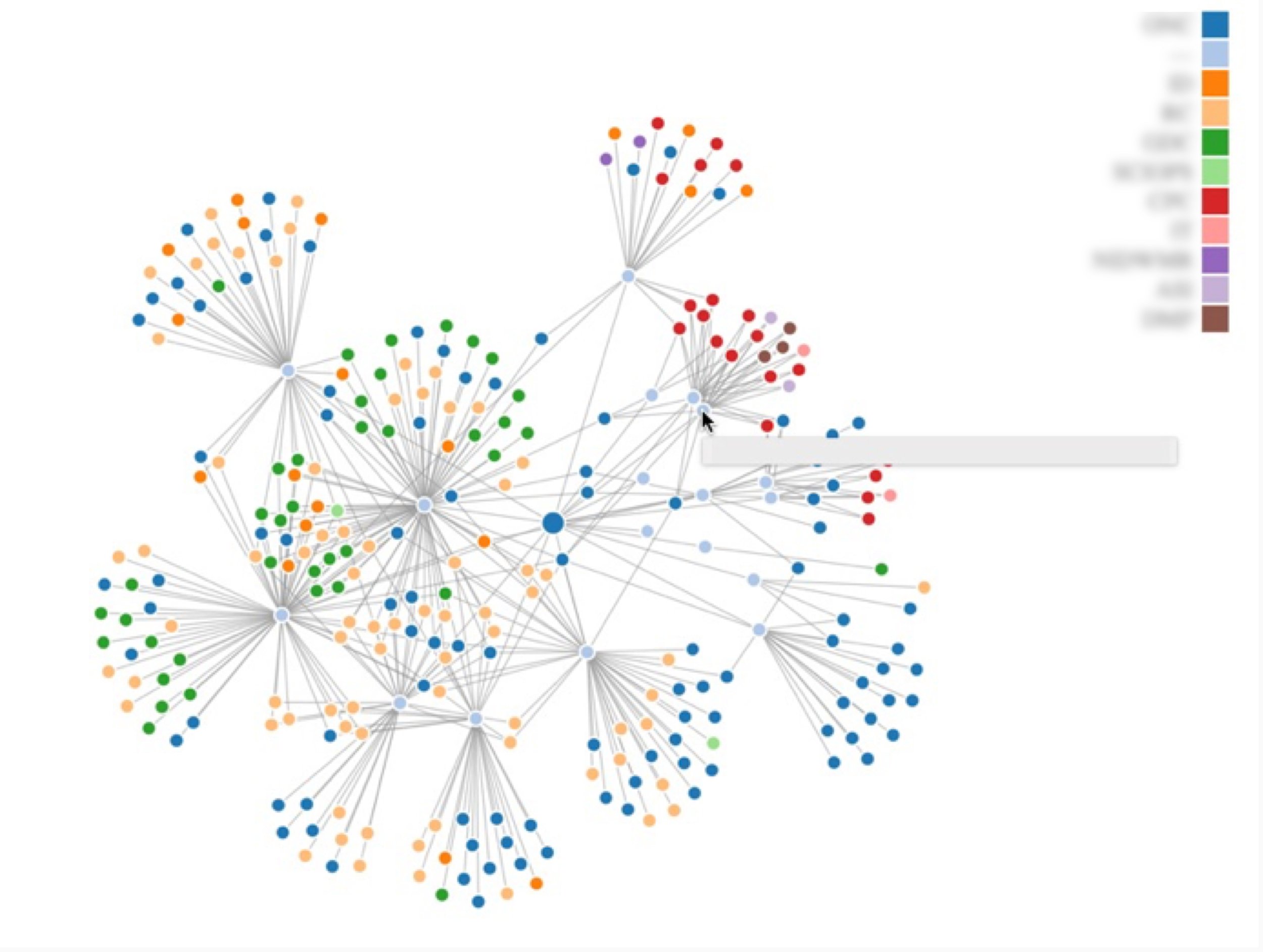
Log analysis: Additionally, I conducted a log analysis to map the expertise available across the organization. This involved analyzing data from different internal systems to identify the types of expertise scientists had and where it was housed. The analysis revealed that the organization had a wealth of expertise, much of it siloed.
Expertise Visualization: I designed a visualization using D3.js to map expertise clusters across the organization. Blurred for confidentiality.
Key Challenges Uncovered:
Organizational silos: Teams were operating in isolated environments with limited cross-functional collaboration. My interviews and workshops revealed that scientists often didn’t have visibility into the work of other teams, hindering their ability to collaborate effectively.
Outdated systems: Surveys and feedback from the workshops made it clear that existing collaboration tools were not intuitive or tailored to the needs of scientists. The log analysis also indicated that archived scientific knowledge was not easily discoverable, leading to duplication of work.
Lack of incentive to share: Interviews with stakeholders and observations during workshops exposed a competitive culture among teams. This reluctance to share knowledge was a significant barrier to progress in drug discovery projects, as each team prioritized their own success over collaborative innovation.
Armed with these insights, I collaborated closely with the product manager and tech leads to develop a roadmap that outlined both short- and long-term design strategies.
Design process
The first solution was the "Activity Stream" app—a feed-based platform to centralize updates on lab activities, instruments, and publications. I began by sketching and developing a series of content wireframes to explore how the activity stream could effectively accommodate the range of content it needed to support.
My next step was to prototype and test with lab scientists. Through iterative user testing, I refined the prototypes based on their feedback, ensuring the design addressed their daily workflow needs. The prototype was built in HTML/CSS/JS, which allowed me to quickly create and test interactive prototypes on different modalities, such as tablet and desktop.
Another project I designed based on this research was the scientist and lab profiles, which centralized key information about individual scientists and labs. This solution aimed to eliminate the need to access multiple sources, making it easier for users to find all relevant data in one place.
Scientist profile prototype: Created with HTML/CSS/JS, blurred for confidentiality.
Collaboration suite app prototype, designed to serve as the team's 'north star,' providing a vision for future development.
Impact
After evaluating the outcomes of this work, our team received recognition from senior leadership for highlighting the critical role collaboration tools play in drug discovery. As a result:
Increased resources: Additional resources were allocated to the product development team to execute the roadmap I had helped shape through research and design.
Strategic leadership alignment: The project was positioned to significantly improve collaboration and efficiency within the drug discovery teams, as discussed in leadership meetings.